Primers and probes can also be quickly tested against large numbers of sequences to see which ones the primers will bind to. Primers can be tested against a single selection on a sequence, against the whole sequence or against multiple entire sequences at once. To test primers, select one or more target sequences then choose Primers → Test with Saved Primers.
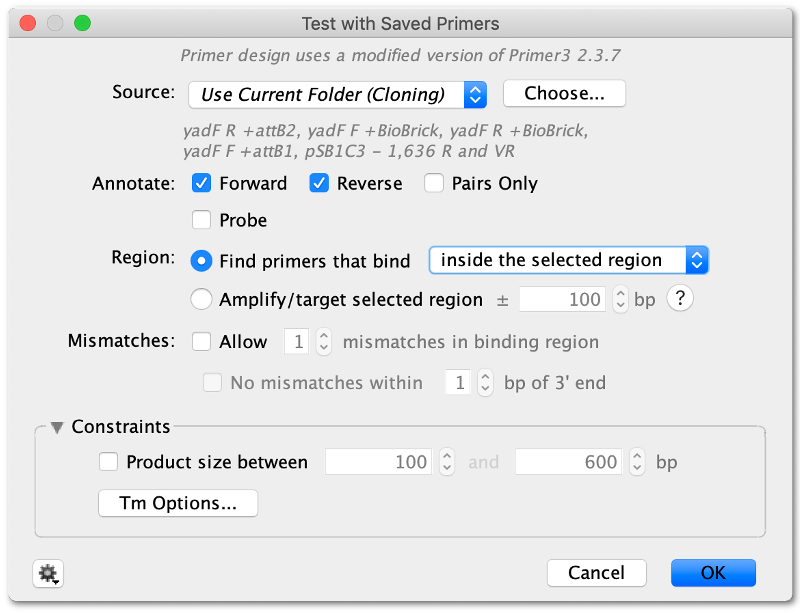
The Source option specifies which primers will be considered for testing on the selected sequence. To test all primers in your local database, choose All Folders. If you select Current Folder, all primers in the same folder(s) as the selected sequence will be tested. Click the Choose button to select any other folder in your local or shared database that contains primers or probes to test.
With Annotate you can limit the primers to a certain direction (relative to the direction of the currently selected sequence). If Pairs Only is selected, inward-directed forward and reverse pairs of primers will be annotated, and the option to limit the Product Size (in the Constraints section) will be enabled. The Pairs Only option normally produces many more annotations, as most primers will be annotated as part of multiple pairs, with all valid partners. If you select the Probe checkbox, oligonucleotide sequences of type “Probe” will be tested (you can create probes when you add a new sequence manually, section 5.1 , or by running “Convert to Oligo”, section 13.8 ).
The Region allows you to choose whether to find primers that bind either anywhere on the sequence or inside the selected region (only enabled if you have selected a region on the sequence you’re testing primers with). Alternatively, if you have selected a region on your sequence, you can select Amplify/target selected region (only available if both Forward and Reverse primers are selected in the Annotate options) to test primers that will amplify the selection, allowing primers to bind upstream or downstream of the selection, as specified. Click the “?” button for more explanation on using the Amplify/target selected region option.
By default, only primers that match the target sequence exactly will be found. If you wish to allow a limited number of mismatches between the primer and target sequence you can specify this under Mismatches, where you can also limit the proximity of mismatches to the 3’ end of the binding site.
Expand the Constraints section in order to limit the Product size range (available if both Forward and Reverse are checked in the Annotate options) or to specify the characteristics used for binding site Tm calculation (by clicking the “Tm Options” button).
Click OK to begin testing primers. Once complete, any primers matching your specified criteria will be annotated on the sequence. If you chose to test the primers as pairs, they will be annotated as pairs on the sequence. The number of matching primers found will be displayed briefly in the status bar below the sequence view.