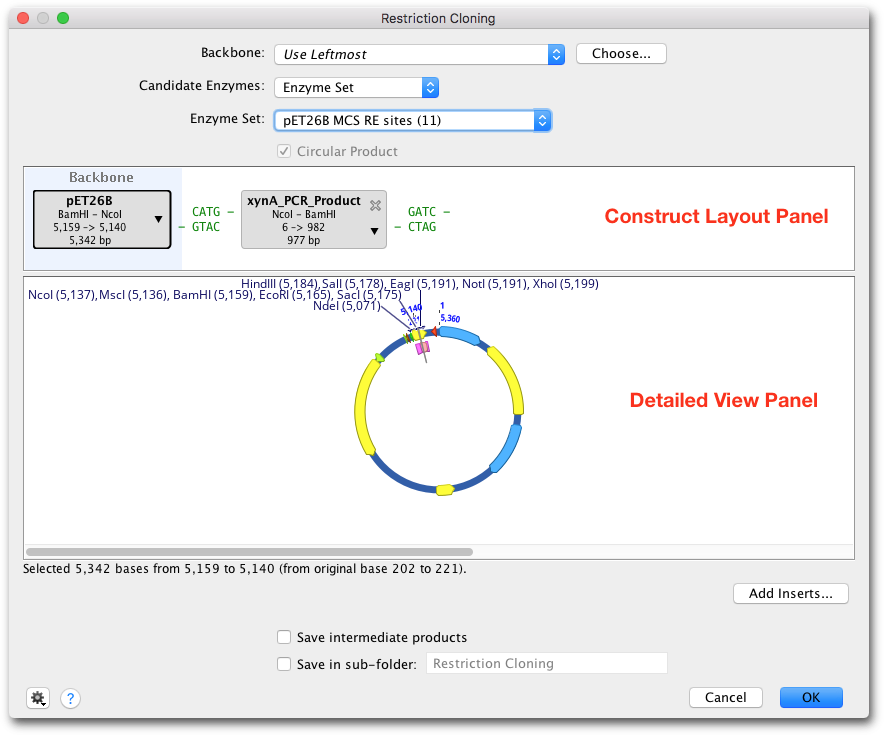
Figure 14.4:
A Restriction Cloning options dialog. The Construct Layout Panel shows the backbone sequence shaded blue to the left and an insert to the right. The Detailed View Panel shows the selected annotated sequence.
Overview
Tools for Restriction Cloning, Golden Gate and Gibson Assembly can be found under the Cloning menu. These tools share a common interface which differs only in options specific to each cloning tool. Input sequences may be selected prior to launching the operation, or may be selected or removed from within the cloning options.
