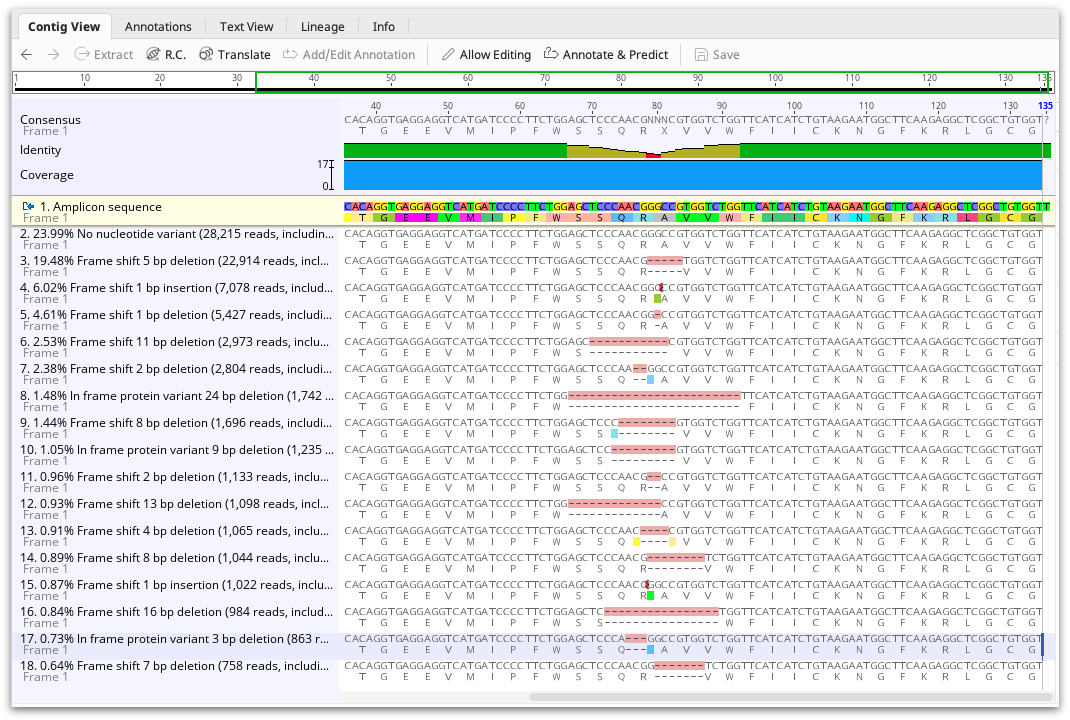
The CRISPR editing analysis tool produces an assembly document showing one representative of each cluster mapped to the reference sequence. The frequency of the variant, and its effect (either No nucleotide variant, Frame shift, In frame stop codon, In frame protein variant, or In frame silent variant) are shown in the sequence name. A summary of these results can also be viewed in the assembly description and the Info tab.
Each variant is also annotated on the mapped read, but annotations are turned off by default. To view the annotations, go to the Annotations and Tracks tab to the right of the viewer and turn on the variant annotations. The annotations contain the same information as the Info tab, such as variant effect and frequency. Using the annotations table, this information can be exported in tabular format.