The colors of nucleotide and amino acid sequences can be set under the General Options tab by clicking the drop down menu next to Colors.
Coloring schemes differ depending on the type of sequence. For example, the Polarity and Hydrophobicity color schemes are available only for protein sequences. Alignments and assemblies can be colored by similarity, read direction and paired distance (if paired reads are used), in addition to standard options.
To change the colors that a particular color scheme uses, click the Edit button then click each base and select the new color you wish to use.
The similarity scheme is used for quickly identifying regions of high similarity in an alignment.
In order for a column to be rendered black (100% similar) all pairs of sites in the column must have a score (according to the specified score matrix) equal to or exceeding the specified threshold.
So for example, if you have a column consisting of only K (Lysine) and R (Arginine) and are using the Blosum62 score matrix with a threshold of 1, then this column will be colored entirely black because the Blosum62 score matrix has a value of 2 for K vs R.
If you raised the threshold to 3, then this column would no longer be considered 100% similar. If the column consisted of 9 K’s and 1 R, then continuing with the threshold value of 3, the 9 K’s which make up 90% of the column would now be colored the dark-grey (80%→100%) range while the single R would remain uncolored.
If instead the column consisted of 7 K’s and 3 R’s (still with threshold 3) then 70% of the column is now similar so those 7 K’s would be colored the lighter grey (60%→80%) range.
Alternatively, going back to the default threshold value of 1, and with a column consisting of 7 K’s, 2 R’s and 1 Y, now since the 7 K’s and 2 R’s have similarity exceeding the threshold whereas the Y is not that similar to K and R, the K’s and R’s will be colored dark grey since they make up 90% of the column.
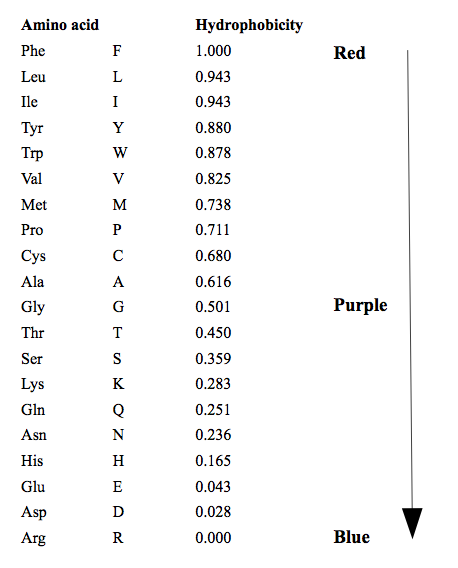
This colors amino acids from red through to blue according to their hydrophobicity value, where red is the most hydrophobic and blue is the most hydrophilic. The values the color scale is based on are given in Figure 5.4 . These values are taken from https://web.expasy.org/protscale/pscale/Hphob.Black.html
This colors amino acids according to their polarity as follows:
Yellow: Non-polar (G, A, V, L, I, F, W, M, P)
Green: Polar, uncharged (S, T, C, Y, N, Q)
Red: Polar, acidic (D, E)
Blue: Polar, basic (K, R, H)