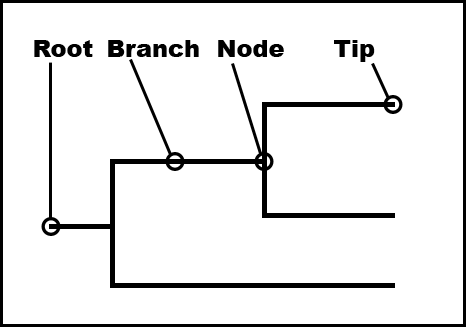
A phylogenetic tree describes the evolutionary relationships amongst a set of sequences. They have a few commonly associated terms that are depicted in Figure 12.1 and are described below.
Branch length. A measure of the amount of divergence between two nodes in the tree. Branch lengths are usually expressed in units of substitutions per site of the sequence alignment.
Nodes or internal nodes of a tree represent the inferred common ancestors of the sequences that are grouped under them.
Tips or leaves of a tree represent the sequences used to construct the tree.
Taxonomic units. These can be species, genes or individuals associated with the tips of the tree.
A phylogenetic tree can be rooted or unrooted. A rooted tree consists of a root, or the common ancestor for all the taxonomic units of the tree. An unrooted tree is one that does not show the position of the root. An unrooted tree can be rooted by adding an outgroup (a species that is distantly related to all the taxonomic units in the tree).
For information on viewing and formatting trees in Geneious, see Section 12.5 .