
Restriction Enzymes1 cut a nucleotide sequence at specific positions relative to the occurrences of the enzyme’s recognition sequence in the sequence. For example, the enzyme EcoRI has the recognition sequence GAATTC and cuts both the strand and the antistrand sequence after the G inside the recognition sequence2 , leaving a single-stranded overhang (sticky end (overhang)):

To find and annotate restriction sites on a nucleotide sequence, go to Find Restriction Sites... under the Tools → Cloning menu or open the Restriction Analysis tab to the right of the sequence viewer.
You can configure the following options:
After configuring your options, click Apply to record the restriction enzyme site annotations on the sequence. The annotation shows the enzyme’s recognition site, cut site and methylation sensitivity. If Highlight Methylation Sites is selected, restriction sites which would not be cleaved due to the presence of methylation by Dam, Dcm or EcoKI are greyed out and marked with an asterisk (for further information on Highlight Methylation Sites see below).
Once the document is saved, two new tabs will appear above the sequence view: Enzymes displays the list of enzymes and their cut positions; Fragments displays a list of fragments that would be produced from the restriction digests. These tables can be exported as .csv files for subsequent processing with other software such as e.g. Microsoft Excel®.
To select the region between two cut sites on a sequence, Shift+click on the two restriction site annotations in the sequence view.
To find enzymes that do not cut a particular sequence, use Find non-cutting enzymes under the Cloning menu. See section 14.3 for further details.
In Geneious Prime 2021 onwards, Geneious will flag restriction sites where restriction enzyme cleavage ability may be affected due to the presence of methyl groups added by methyltransferases Dam, Dcm, and EcoKI.
This is determined by experimental validation according to rebase.neb.com, which uses the following terms to describe the effect of methylation:
|
|
|
| Cut | Not sensitive to methylation at the overlapping site |
| Impaired | Rate of cleavage is lower than for unmethylated overlapping site |
| Blocked | Will not cleave when overlapping site is methylated |
| Some blocked | Blocked by methylation of the overlapping site in some but not all flanking DNA contexts |
| Variable | Conflicting reports of sensitivity to methylation at the overlapping site |
| Untested | Effects of methylation at the overlapping site have not been tested |
|
|
|
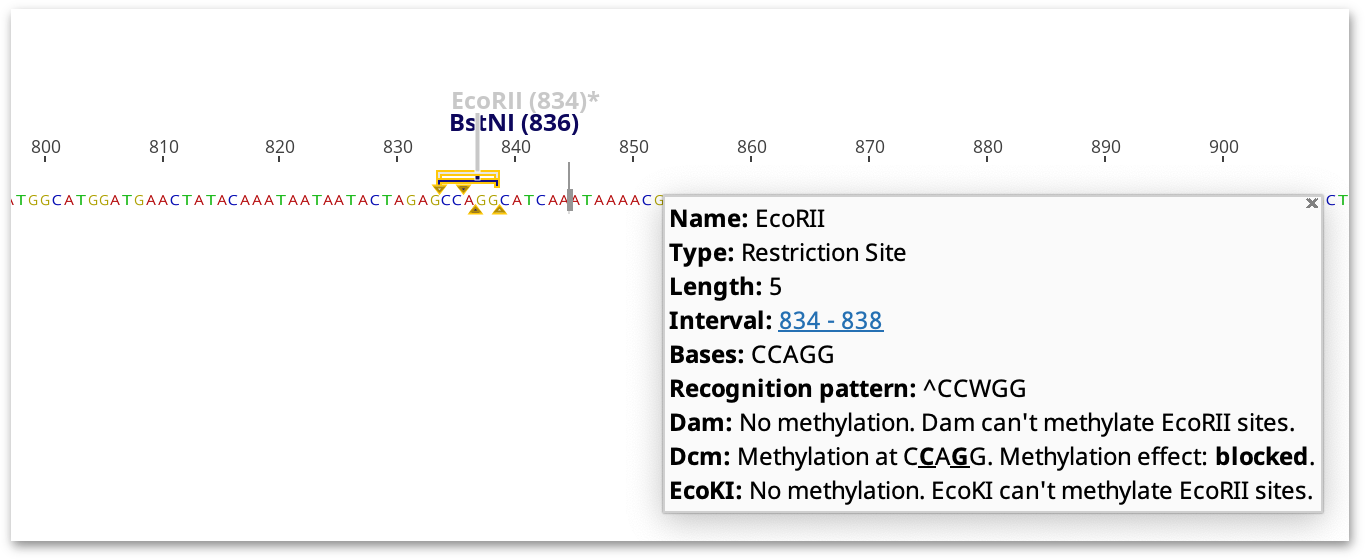
Restriction sites are greyed out and marked with an asterisk if the ability of the restriction enzyme to cleave at the restriction site is variable, impaired or blocked (see Figure 14.1 ). Methylated restriction sites where cleavage has not been experimentally validated, or might be methylated but have ambiguities in the target sequence, are highlighted with an asterisk but not greyed out.
The highlighted restriction sites and indicated methylation effect takes into consideration the neighboring sequence. For example, cleavage by XbaI is only blocked by dcm methylation if the recognition site (TCTAGA) is preceded by GA or followed by TC. If these flanking bases are not present, the site will not be marked as affected by methylation, but a note on the restriction site annotation will say what the effect of methylation would be if the correct bases were present.
Effective length for restriction enzymes is displayed in both the Advanced table of enzymes, and in the Enzymes tab on the sequence viewer.
Effective length is a measure of how frequently an enzyme will cut, taking into account both sequence length and ambiguities. In other words, lower effective length means an enzyme is expected to cut more frequently. Because ambiguous bases are more likely to match a sequence by chance, they contribute less than 1 to the effective length.
Effective length is calculated as the sum of the following formula across all symbols in the recognition sequence, where n is the number of nucleotides each symbol represents.
1 - log(n) / log(4)
I.e. 1 for a ACGT, 0.5 for 2-ambiguity (MRWSYK), .208s for 3-ambiguity (VHDB) and 0 for N.
Note: The sum displayed in Geneious is rounded down to nearest .0 or .5
See http://search.cpan.org/dist/BioPerl/Bio/Restriction/Enzyme.pm#cutter for a little explanation of why you would use this.