The Virtual Gel tab will appear above the sequence viewer when nucleotide documents containing fewer than 10,000 sequences or 1000 restriction sites are selected.
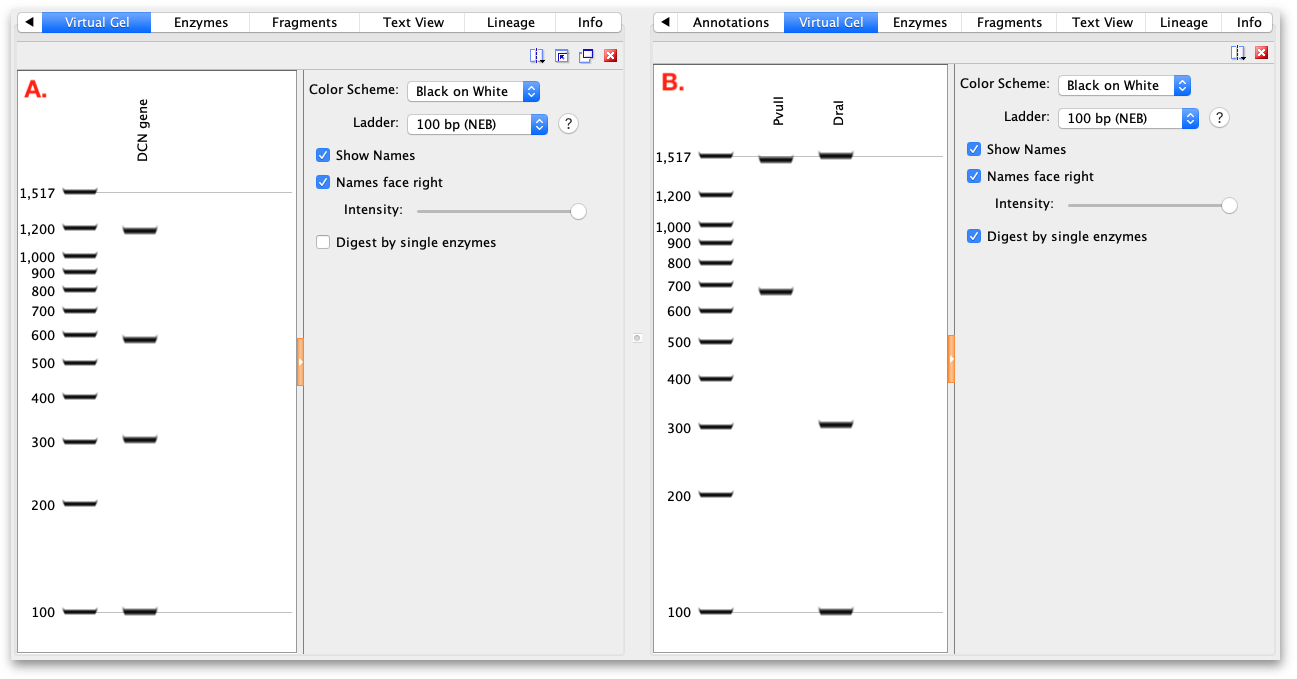
If a single sequence annotated with restriction enzyme sites is selected, the gel displays the fragments that would result from that restriction digest. If the restriction sites are all from a single enzyme, the digest pattern is shown in one lane of the gel.
If multiple different restriction enzymes are annotated, the results of digestion with each enzyme will be shown in different lanes if Digest by single enzymes is checked. If this option is unchecked, the result will be shown as a multiple enzyme digest in a single lane (see Figure 5.7 )

If multiple sequence documents containing restriction sites are selected (either by bulk-selecting individual sequence documents, or selecting a sequence list), digests for each sequence are shown in separate lanes on the gel.
If the sequence list contains sequences with no annotated restriction sites (for example the results of the Digest into Fragments operation) then all sequences will be displayed in one lane of the virtual gel.
By default, sequences will appear on the gel in the order they are displayed in the document table or sequence list. To change the order of display, simply drag and drop the sequences on the gel to the desired position.