15.1 Setting up a BLAST search
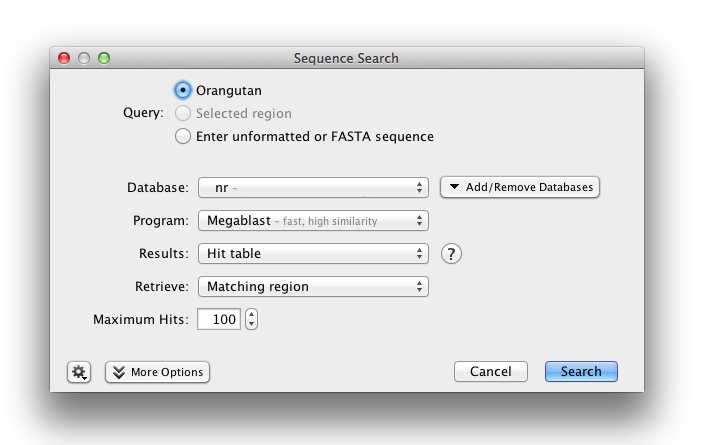
To run a BLAST search in Geneious Prime, select your query sequence or sequences and click the BLAST button in the toolbar. This operation can also be accessed by going to the Tools menu or by right-clicking (Ctrl+click on Mac OS X) on a sequence document and choosing BLAST. You can choose to BLAST either your currently selected sequence documents or a sequence you enter manually. If you choose to enter your sequence manually, then Geneious will display a large text box in which you can enter your query sequence as either unformatted text or FASTA format.
Select your database using the first drop-down box. Databases are grouped together under their respective services. Then choose which kind of BLAST search you wish to run under Program. The available programs will depend on the database you have chosen.
Geneious Prime can perform seven different kinds of BLAST search:
- blastn: Compares a nucleotide query sequence against a nucleotide sequence database.
- Megablast: A variation on blastn that is faster but only finds matches with high similarity.
- Discontiguous Megablast: A variation on blastn that is slower but more sensitive. It will find more dissimilar matches so it is ideal for cross-species comparison.
- blastp: Compares an amino acid query sequence against a protein sequence database.
- blastx: Compares a nucleotide query sequence translated in all reading frames against a protein sequence database. You could use this option to find potential translation products of an unknown nucleotide sequence.
- tblastn: Compares a protein query sequence against a nucleotide sequence database dynamically translated in all reading frames.
- tblastx: Compares the six-frame translations of a nucleotide query sequence against the six-frame translations of a nucleotide sequence database.
Three options are available for displaying your results:
- Hit table: Returns one alignment for every hit against the database and displays them in a hit table. Each query displays a separate table and is also viewable as a query-centric alignment. This is suitable for less than 100 queries.
- Query-centric alignment: Returns one alignment for each query, showing the hits aligned against the query sequence. This is well suited for large batch searches but it doesn’t display a hit table.
- Bin into ’has hit’ vs. ’no hit’ Returns two sequence lists: one containing queries which get a hit in the database, the other containing queries which don’t. Details about the hits and alignments are discarded. This can be used to filter contamination (eg. human) from sequencing reads.
You can also specify how much of each matching sequence to retrieve from your database:
- Matching region: Just the region of the database sequence which matches the query.
- Matching region with annotations: The region of the database sequence which matches the query, plus any annotations on that sequence.
- Extended region with annotations: The matching region plus additional flanking regions upstream and downstream.
- Full sequence with annotations: The entire database sequence (this could be large and slow).
Geneious also allows you to specify most of the advanced options that are available in BLAST. To access the advanced options click the More Options button which is in the bottom left of the BLAST options. The available options vary depending on the kind of BLAST search you have selected. For details on each of the options you can hover your mouse over the option to see a short description or refer to the BLAST documentation from NCBI.