14.2 Digest into fragments
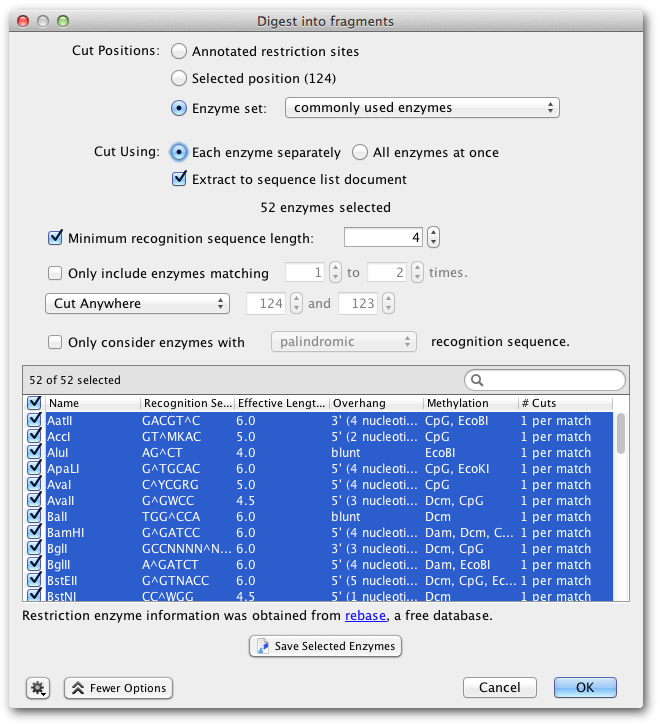
The option Digest into fragments... from the Tools → Cloning menu or the context menu allows you to generate the nucleotide sequences that would result from a digestion experiment. You can digest multiple nucleotide sequences at a time. If the digestion results in overhangs, these will be recorded as annotations on the fragments.
- If you have selected only one nucleotide sequence document and it has annotated restriction sites, you can select Annotated cut positions to cut the document on these sites. When this option is selected, the options to filter the enzymes by their effective recognition sequence length or number of hits are disabled. You can select a subset of the annotated enzyme sites under More Options.
- If you do not have annotated cut sites already on your document, you can choose Enzyme set and select which enzyme(s) you wish to use. This runs Find Restriction Sites first, but does not generate restriction site annotations on your original sequence. See the section on Find Restriction Sites (14.1
) for more detail.
- Where multiple enzymes are selected, you can either digest the original sequence by Each enzyme separately, which returns a separate sequence list of the fragments produced for each enzyme, or by All enzymes at once, which digests by all selected enzymes in one operation.