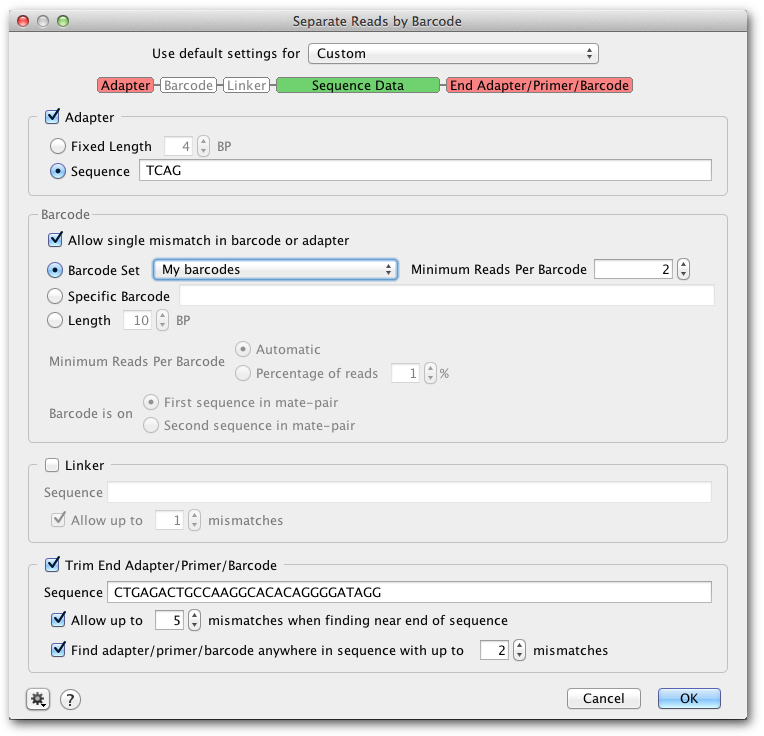
Multiplex or barcode data (e.g. 454 MID data) can be separated using Separate Reads by Barcode from the Sequence menu. This function copies all sequences matching a given barcode to a correspondingly named sequence list document.

Default settings are provided for 454 standard and Titanium MID barcodes (with or without Adaptor B trimming), and Rapid MID barcodes. These settings recognise standard MID sequences provided by 454 and use their names when appropriate.
To enter a custom barcode set, select Custom settings, then under Barcode set choose Edit barcode sets. Then click Add to add your list of barcodes. To specify fixed sequences either side of the barcode, enter these in the Adaptor and Linker sections.
If you only want to extract sequences with a single, specific barcode sequence (e.g. a primer), check Specific barcode and enter the sequence. Alternatively if you do not know your barcode sequences, you can just enter the length of your barcodes and Geneious will automatically identify what the barcodes are.
Separate reads by barcodes only sorts by barcodes at the 5‘ end of the sequence, but primers, adapters or barcodes on the 3‘ end of the sequence can be trimmed off by checking Trim End Adaptor/Primer/Barcode. Either enter a specific sequence to trim, or add your 3‘ barcode to your custom barcode set and use the text [END_BARCODE] in the sequence box. Primer trimming can also be performed after separating by barcodes using Trim Ends (see section 10.2.2 ).
For further information on splitting barcode data, hover the mouse over any of the settings in the Separate Reads by Barcode options window.