Geneious Prime can compare annotations across up to 3 annotation tracks or documents, highlighting annotations which are common or unique depending on which criteria you choose.
To use this function, select the sequence or sequences containing annotations you wish to compare, and go to Annotate and Predict → Compare Annotations. In the Annotation Types panel, select the annotations you wish to compare. The default setting is for pairwise comparison; if you wish to do three-way comparison select Set C. Choose the annotation type and location for each set, and the type of comparison.
Comparison options are:
A new annotation track will be created showing the results of the comparison. The results panel in the compare annotations set up allows you to choose which comparison to return:
More than one of these options can be selected at once, either by checking the box next to the option, or clicking the appropriate section of the venn diagram to the right. Each result comparison is displayed on a separate track on the original sequence, and a preview of these tracks is given in the Example panel.
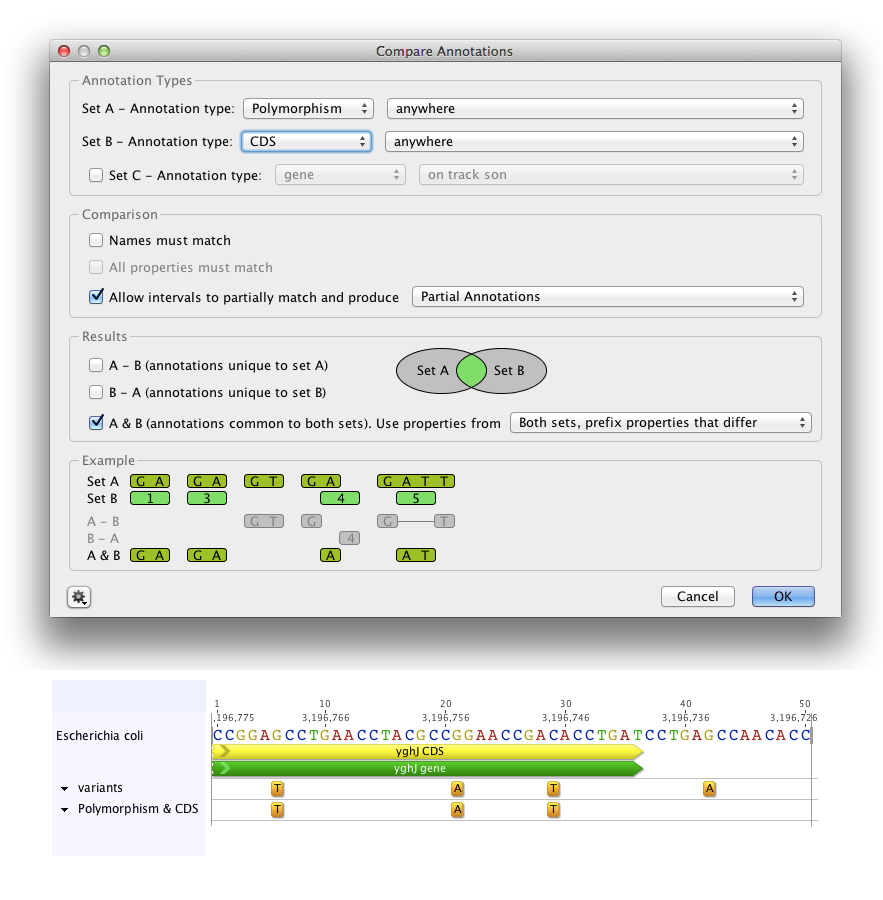
To return a track containing polymorphism annotations that are within another gene or feature, such as a coding sequence, select the Polymorphism annotation type for Set A, and choose which track it is on (or select anywhere if there are no tracks, or you want to include polymorphisms on any track). Then choose the other annotation type (e.g. CDS) for Set B. Uncheck Names must match as in general polymorphism annotation names do not match those of other annotation types, and check Allow intervals to partially match and produce partial annotations. Under Results, choose to return A & B (annotations common to both sets). This will return a new track containing annotations of the type in Set A (polymorphisms) that are contained within Set B (CDS) annotations. See Figure 8.6 .
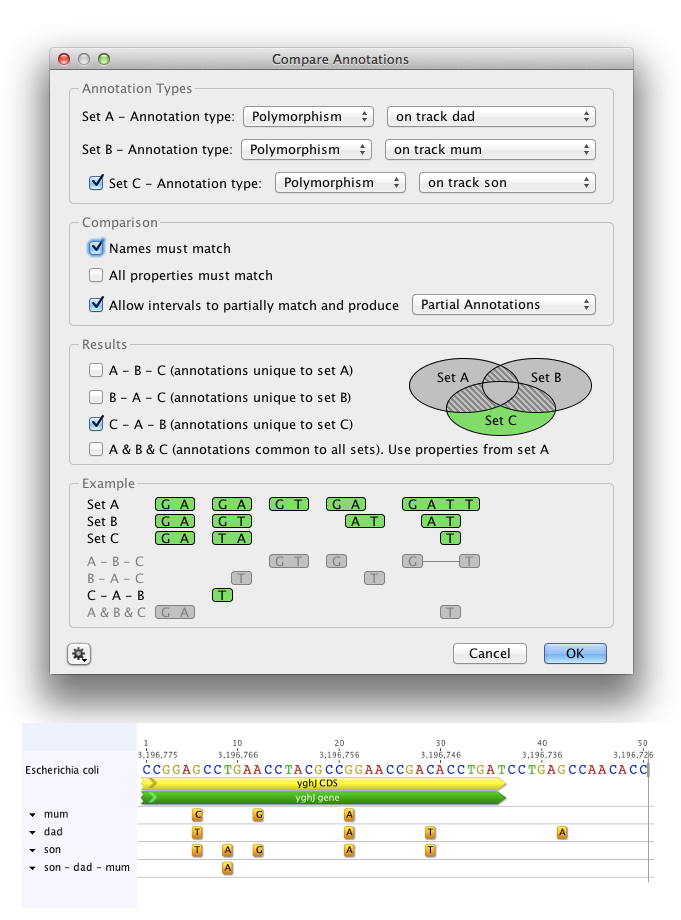
To return polymorphisms which are unique to the offspring, set Annotation Type as Polymorphism for Sets A, B and C, and set A and B as the parent tracks, and C as the child track, as in Figure 8.7 . Check Names must match and Allow intervals to partially match and produce partial annotations. Choose C-A-B in the results display. This will return a new track containing annotations found in Set C (child), but not in Sets A and B (parents).